A very brief intro to proteins
Dr.Mikko Karttunen
Department of Chemistry
Waterloo Institute for Nanotechnology University of Waterloo, Ontario, Canada
Web: www.softsimu.org Email: mkarttu@gmail.com
Protein structure-function paradigm
Figure: Based on Mariana Ruiz Villarreal's illustration on Wikipedia
More on protein structure: [Nature Education] [Molecular Biology of the Cell] [Wikipedia] [Amino acids]
Protein structure Nobel Prizes: [Chemistry 1946] [Chemistry 1962] [Chemistry 1964] [Chemistry 1972] [Chemistry 2002]
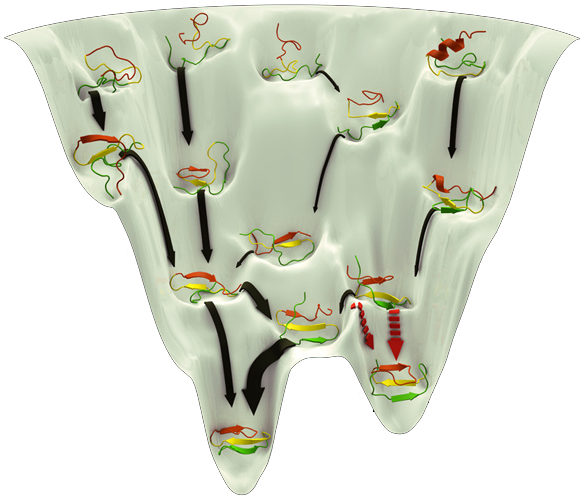
Globular protein: Ubiquitin
PDB structure: 1UBQ [Folding Funnel Diagrams: M. Karplus, Nat. Chem. Biol 7, 401 (2011)]
More on Ubiquitin: [Molecule of the month] [UBQ on Wikipedia] [Nobel Prize in Chemistry 2004]
"Intrinsically disordered proteins (IDP) are different
Structural ensembles: IDP vs a 'traditional' protein.
IDPs vs globular proteins
Typical amino acid compositions are different.
- IDPs have more charged residues
- more polar residues
- more structure breaking residues (glycine & proline)
- less aromatic content
- less hydrophobic content
One reason for the dynamic behavior:
- Lack of hydrophobic cores makes them more dynamic
Structure:
- Lack of tertiary structure
- There are elements of secondary structure
- Linear motifs. Crucial for binding
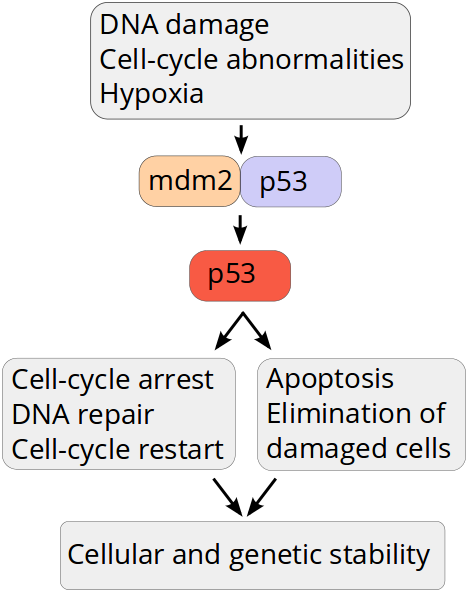
p53 - "the Guardian of Genome"
"Conformational biases of linear motifs",
E. Cino, W.-Y. Choy, M. Karttunen,
J. Phys. Chem. B 117, 15943 (2013).
Some general properties IDPs have
Advatages IDPs have:
- Specificity without excessive binding strength
- Fast, switch-like, binding
- One-to-many signalling: Protein hubs
Hard to get experimental data:
- There is in vitro evidence
- There are in vivo hypotheses
- Computational methods offer an alternative
Diseases:
- Signalling related
- Often cancer-related
- Oxidative stress
- Diabetes 2
- Neurodegenerative diseases
p53, "the Guardian of Genome"
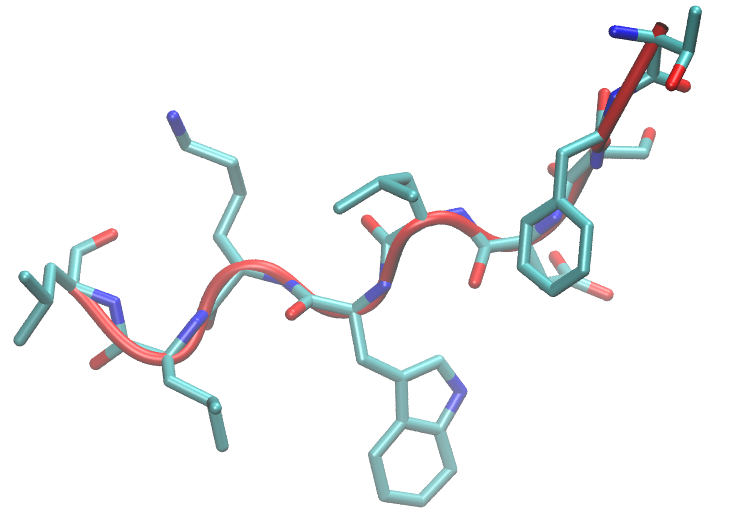
"Comparison of secondary structure formation using 10 different force fields in microsecond molecular dynamics simulations", Cino, Choy, Karttunen, J. Chem. Theory and Comput. 8, 2725 (2012)
Comparison of 10 different force fields
Comparison of secondary structure formation using 10 different force fields
in microsecond molecular dynamics simulations,
Cino, Choy, Karttunen,
J. Chem. Theory and Comput. 8, 2725-2740 (2012)
Binding of disordered proteins to a protein hub,
Cino, Killoran, Karttunen, Choy,
Sci. Rep. 3, 2305 (2013).
Intrinsically Disordered Protein: Exoenzyme S
- Conformational biases of linear motifs, J. Phys. Chem. B 117, 15943–15957 (2013)
- Binding of disordered proteins to a protein hub, Scientific Reports 3, 2305 (2013).
- Effects of Molecular Crowding on the Dynamics of IDPs, PLoS ONE 7 e49876 (2012).
- Comparison of secondary structure formation using 10 different force fields, JCTC 8, 2725-2740 (2012)
- Microsecond MD Simulations of IDPs Involved in the Oxidative Stress, PLoS One 6, e27371 (2011).